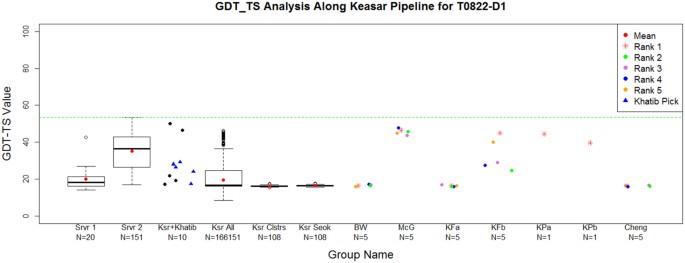
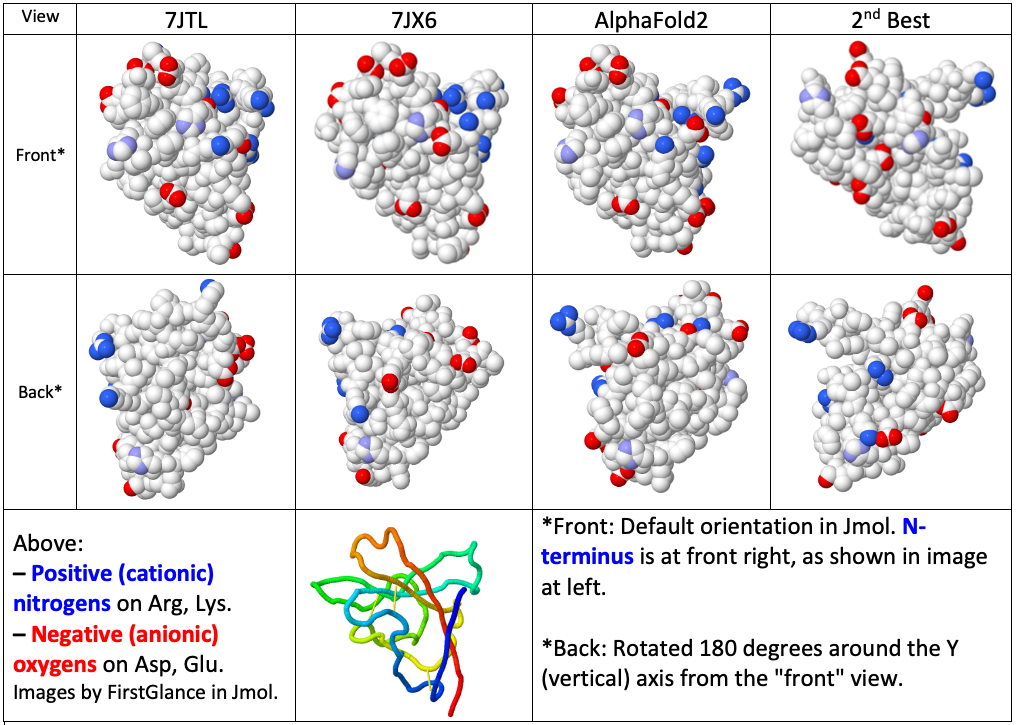
Topology evaluation of models for difficult targets in the 14th round of the critical assessment of protein structure prediction (CASP14) - Kinch - 2021 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
An analysis and evaluation of the WeFold collaborative for protein structure prediction and its pipelines in CASP11 and CASP12 | Scientific Reports

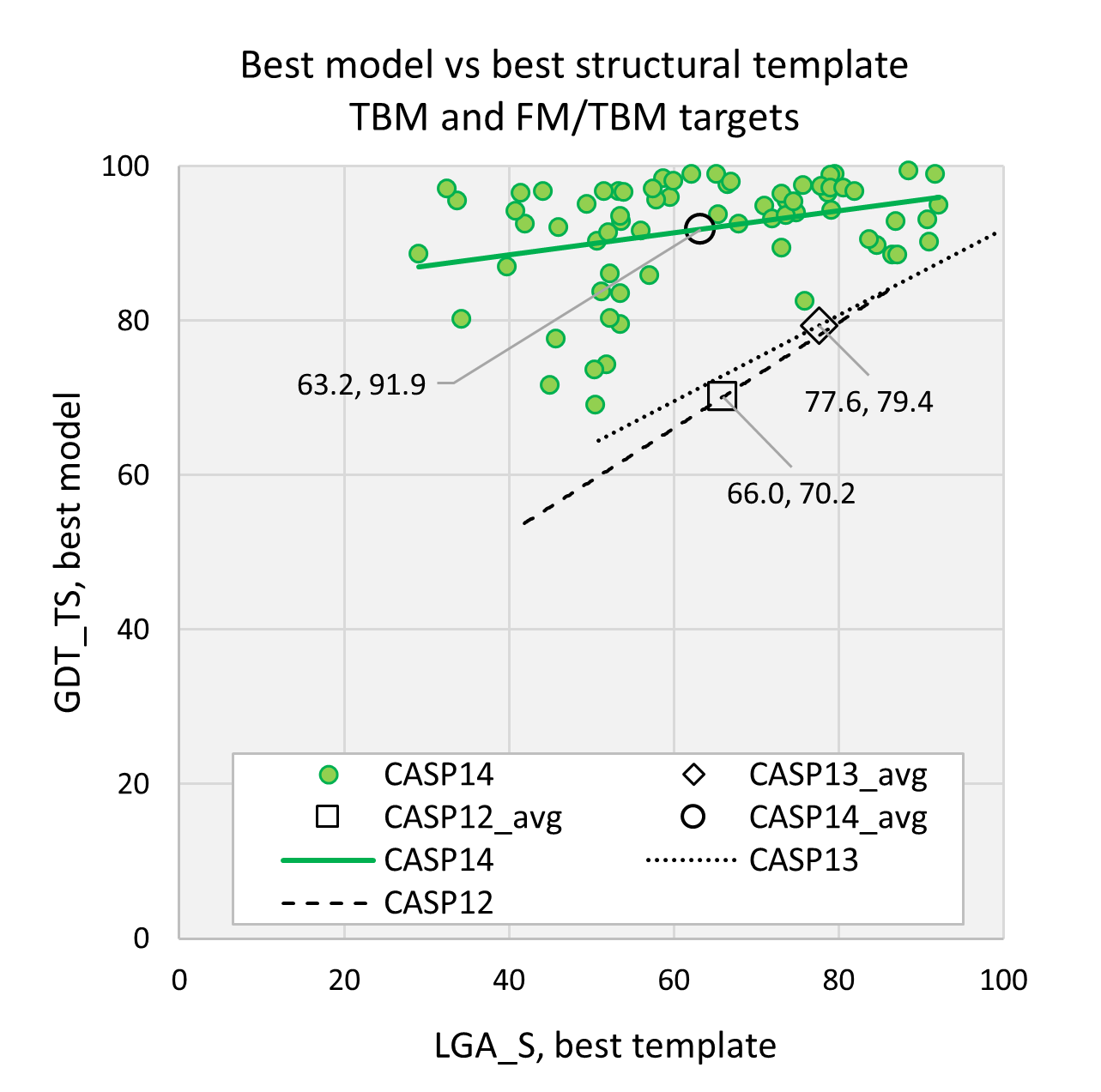
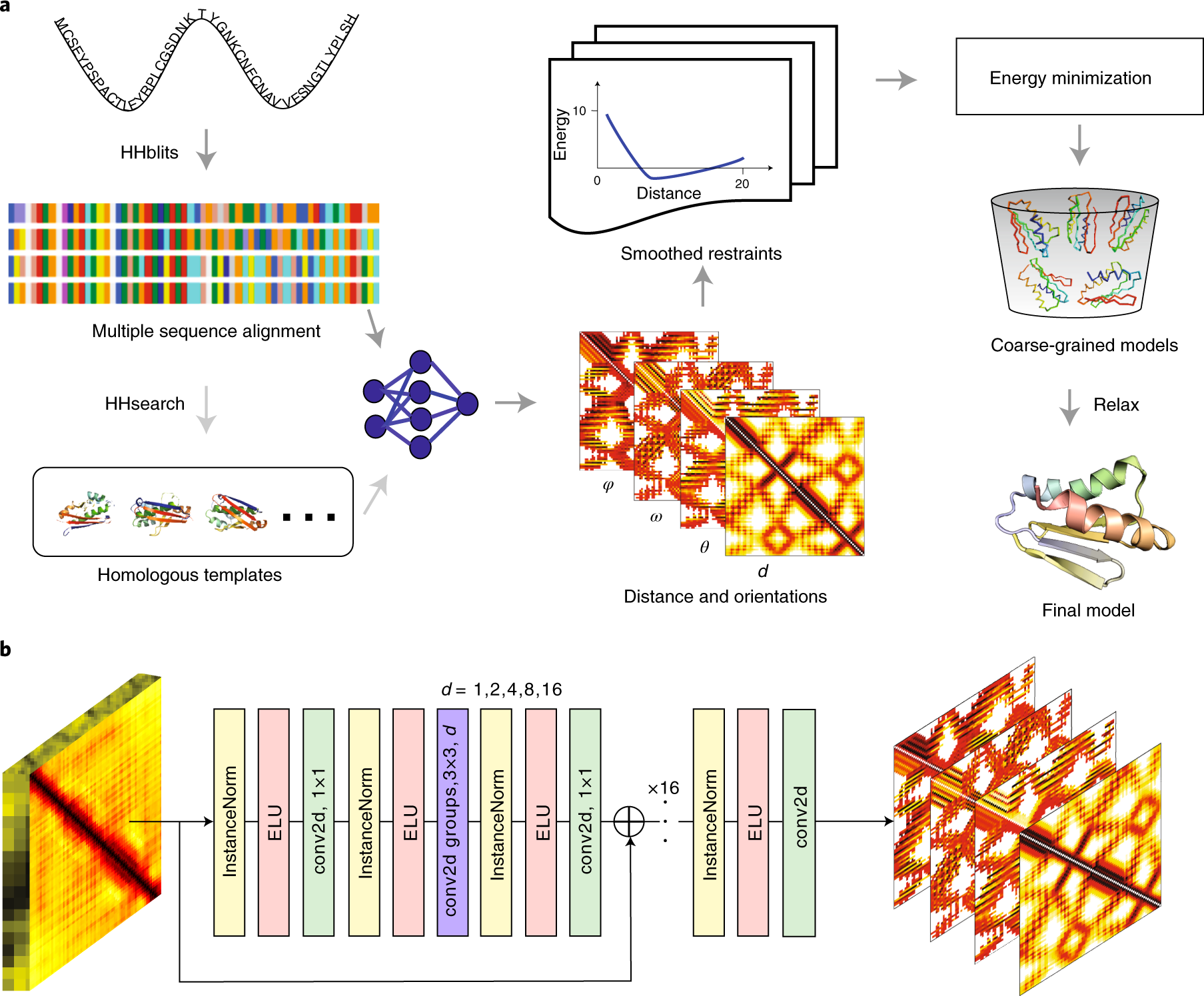
Improving protein tertiary structure prediction by deep learning and distance prediction in CASP14 - Liu - 2022 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library

ROPIUS0: A deep learning-based protocol for protein structure prediction and model selection and its performance in CASP14 | bioRxiv

Topology evaluation of models for difficult targets in the 14th round of the critical assessment of protein structure prediction (CASP14) - Kinch - 2021 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
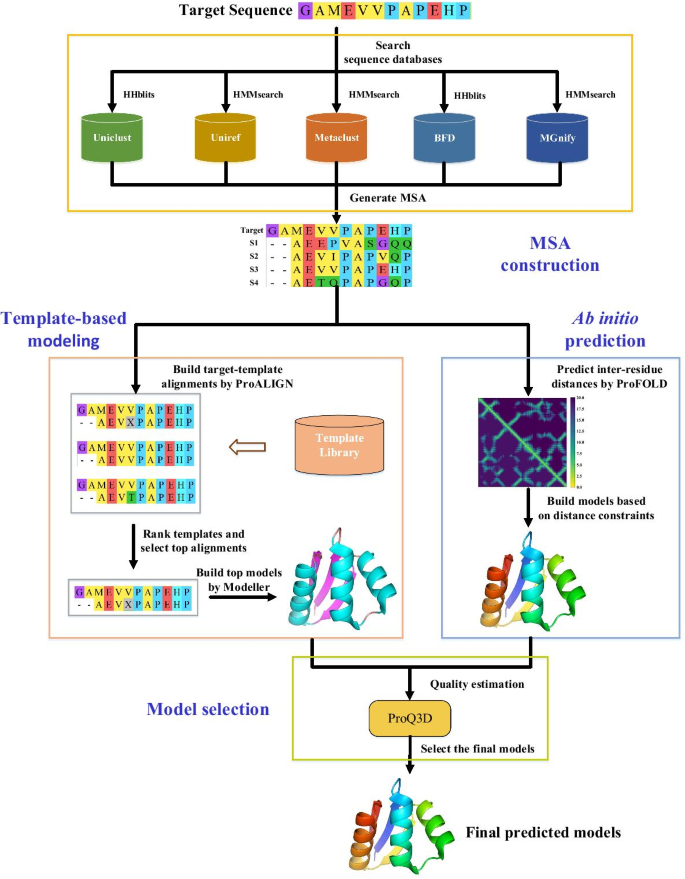
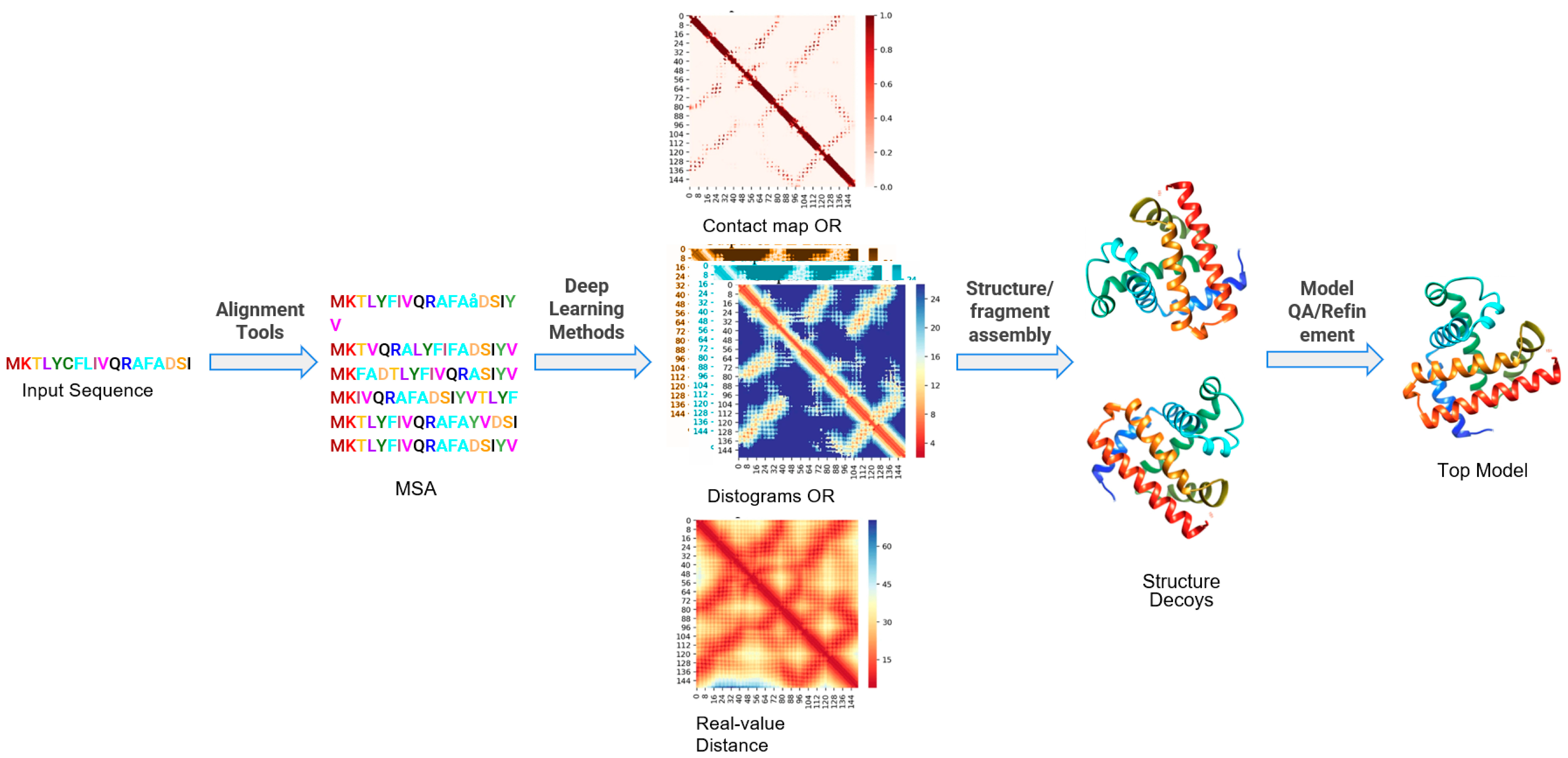
FALCON2: a web server for high-quality prediction of protein tertiary structures | BMC Bioinformatics | Full Text

Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13 | bioRxiv

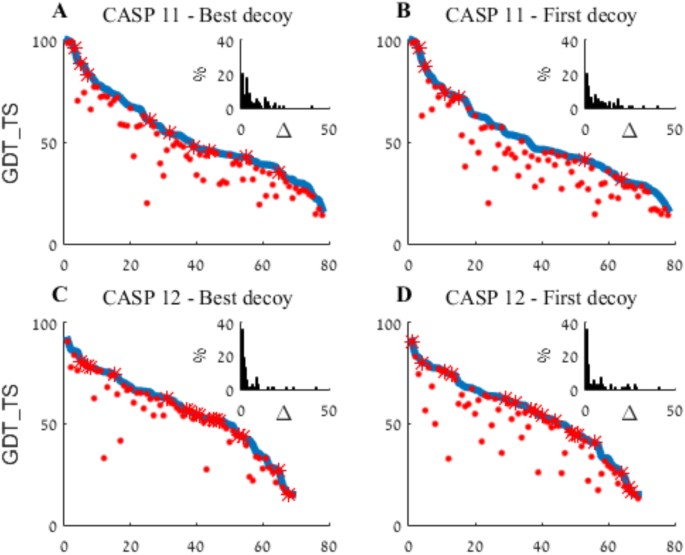
Assessment of protein model structure accuracy estimation in CASP13: Challenges in the era of deep learning - Won - 2019 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library

CASP14: what Google DeepMind's AlphaFold 2 really achieved, and what it means for protein folding, biology and bioinformatics | Oxford Protein Informatics Group

Assessment of protein model structure accuracy estimation in CASP14: Old and new challenges - Kwon - 2021 - Proteins: Structure, Function, and Bioinformatics - Wiley Online Library
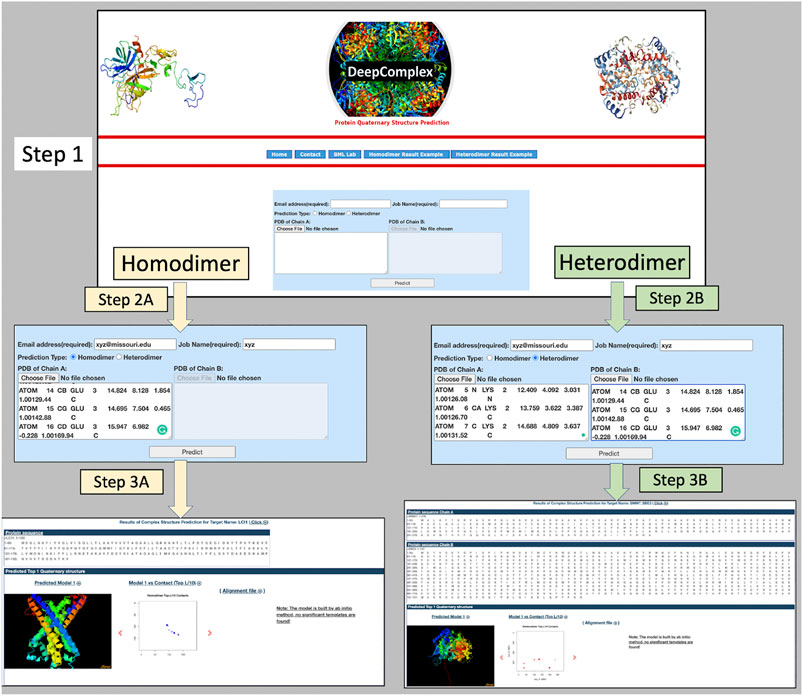
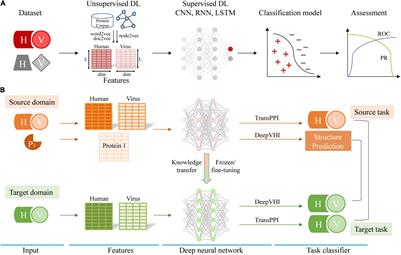
Frontiers | DeepComplex: A Web Server of Predicting Protein Complex Structures by Deep Learning Inter-chain Contact Prediction and Distance-Based Modelling