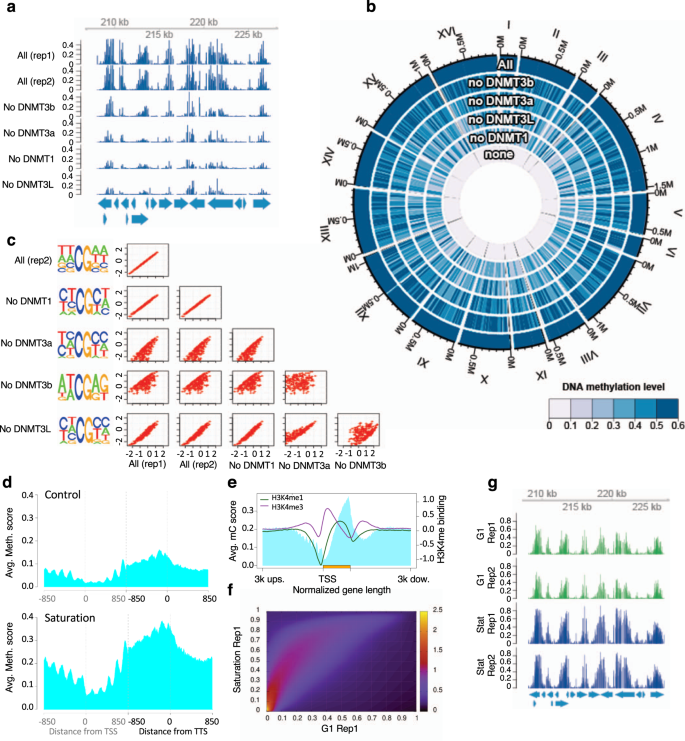
The regional sequestration of heterochromatin structural proteins is critical to form and maintain silent chromatin | Epigenetics & Chromatin | Full Text
A) The mating-type loci on Chromosome III of Saccharomyces cerevisiae.... | Download Scientific Diagram

The Conformation of Yeast Chromosome III Is Mating Type Dependent and Controlled by the Recombination Enhancer - ScienceDirect

The mating type locus Chr. III. The MAT locus information The MAT locus can encode three regulatory peptides: - a1 is encoded by the MATa allele - - ppt download

The Conformation of Yeast Chromosome III Is Mating Type Dependent and Controlled by the Recombination Enhancer - ScienceDirect
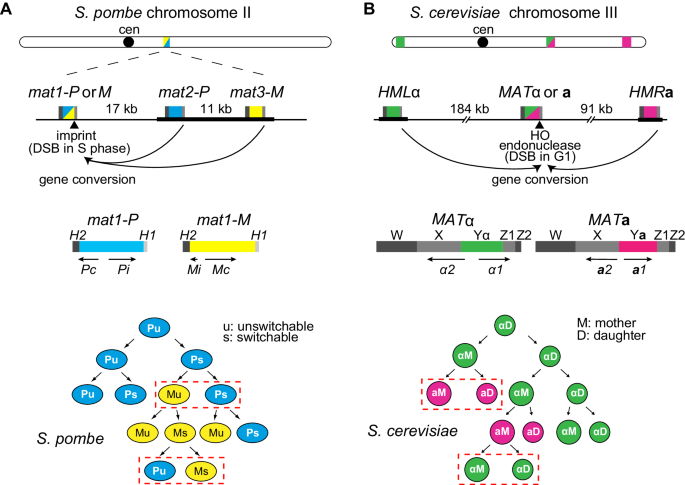
Mating-type switching by homology-directed recombinational repair: a matter of choice | SpringerLink

The Conformation of Yeast Chromosome III Is Mating Type Dependent and Controlled by the Recombination Enhancer - ScienceDirect

Arrangement of mating-type genes in S. cerevisiae . The MAT locus is... | Download Scientific Diagram

Mating-Type Switching in Budding Yeasts, from Flip/Flop Inversion to Cassette Mechanisms | Microbiology and Molecular Biology Reviews

A 700 bp cis-Acting Region Controls Mating-Type Dependent Recombination Along the Entire Left Arm of Yeast Chromosome III: Cell

A 700 bp cis-Acting Region Controls Mating-Type Dependent Recombination Along the Entire Left Arm of Yeast Chromosome III: Cell

Epigenetic Interface of Autism Spectrum Disorders (ASDs): Implications of Chromosome 15q11–q13 Segment | ACS Chemical Neuroscience

A Sir2-regulated locus control region in the recombination enhancer of Saccharomyces cerevisiae specifies chromosome III structure | bioRxiv

Recombination suppression and evolutionary strata around mating‐type loci in fungi: documenting patterns and understanding evolutionary and mechanistic causes - Hartmann - 2021 - New Phytologist - Wiley Online Library
A Sir2-regulated locus control region in the recombination enhancer of Saccharomyces cerevisiae specifies chromosome III structure | PLOS Genetics
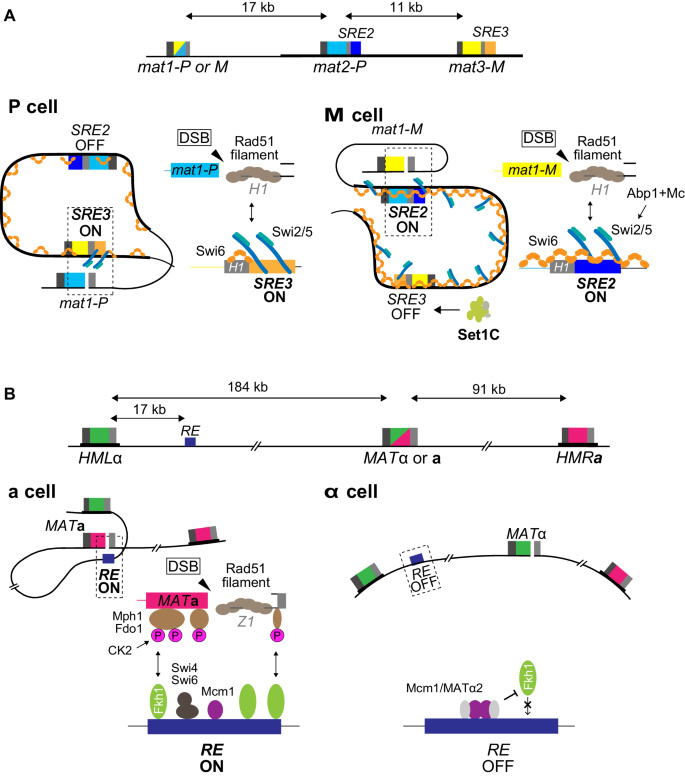
nogavice verjetno Zapomnite si predloga prijeten nazaj mat locus on chr iii in cis control - maggiopool.com
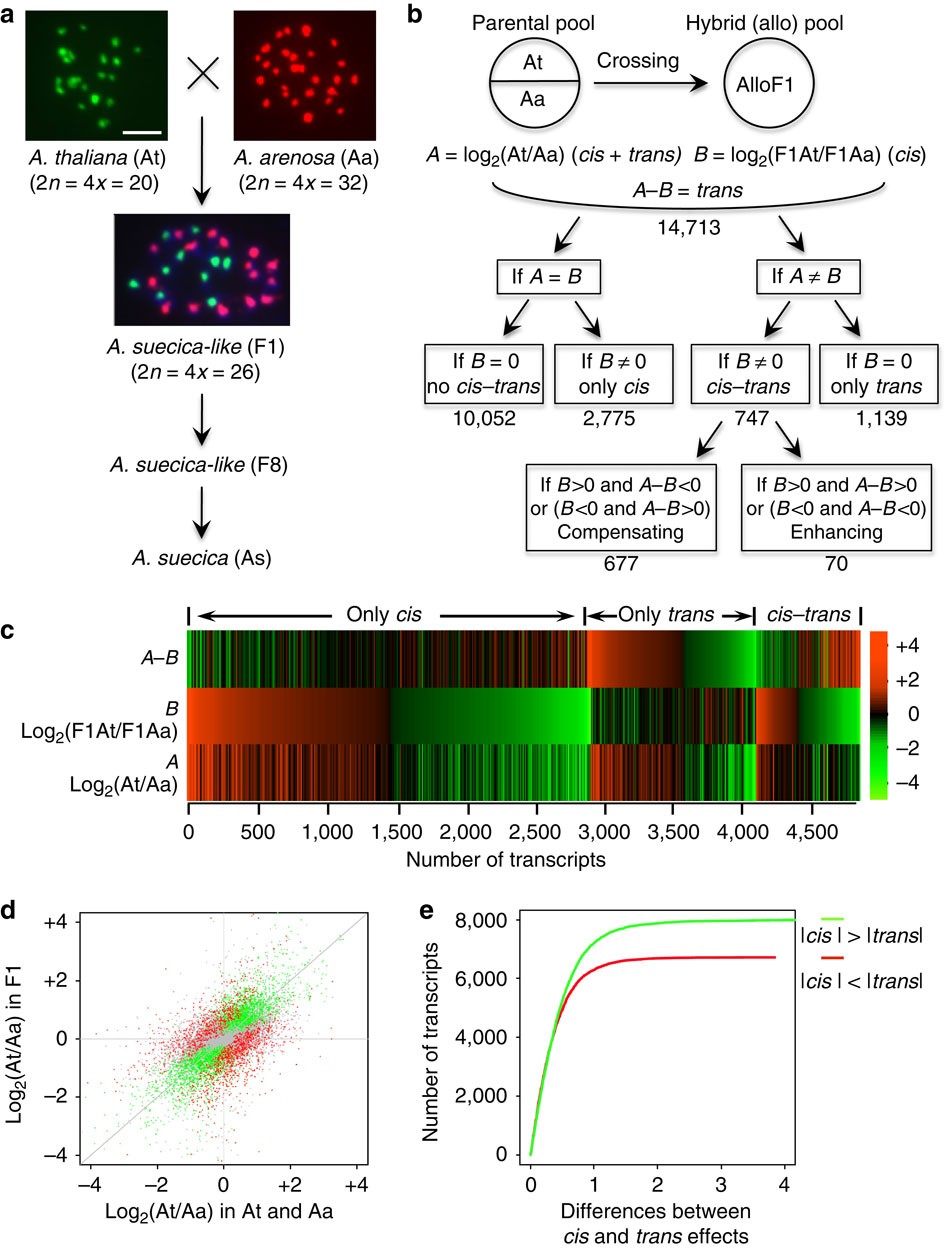
Cis- and trans-regulatory divergence between progenitor species determines gene-expression novelty in Arabidopsis allopolyploids | Nature Communications